Discovery of antimicrobial peptides in the global microbiome with machine learning
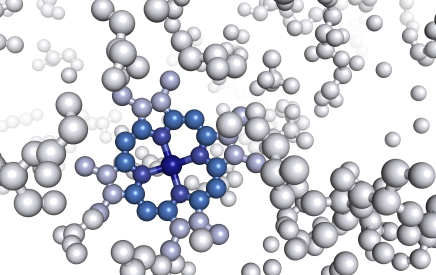
A machine-learning-based approach predicts antimicrobial peptides (AMPs) within the global microbiome using a vast dataset of 63,410 metagenomes and 87,920 prokaryotic genomes. The AMPSphere catalog contains 863,498 non-redundant peptides, providing insights into their evolutionary origins. The approach validates predictions by synthesizing and testing 100 AMPs against drug-resistant pathogens and human gut commensals. 79 active AMPs were identified, with 63 targeting pathogens and disrupting bacterial membranes. This approach provides an open-access resource for antibiotic discovery.
AMR NEWS
Your Biweekly Source for Global AMR Insights!
Stay informed with the essential newsletter that brings together all the latest One Health news on antimicrobial resistance. Delivered straight to your inbox every two weeks, AMR NEWS provides a curated selection of international insights, key publications, and the latest updates in the fight against AMR.
Don’t miss out on staying ahead in the global AMR movement—subscribe now!